Current genetics
Multimedia E-textbook of medical biology, genetics and genomicsGlossary of genetic terms
This section of Current Gentics should, at least according to our original intention, in fact arise gradually from the interaction of students (and possibly other users) and teachers/authors. Repeatedly, some genetic terms are brought up for clarification by students in class or during consulting session. Although some of the included terms would deserve an individual chapter (and they might eventually even end up in this form), at this site we would like to gather short summaries and defintions of the terms most frequently appearing in YOUR queries, given they fall into the broad field of genetics and genomics. Please post your inquiries and suggestions to the e-mail address available at the bottom of the page. Thank you for your input and we hope that the Glossary will fullfil its planned role.
Ames test
Ames test is used for identification of mutagenic substances. In 60's of 20th century Bruce Ames developed a test that still bears his name and is still used as a fast and relatively cheap way to assess the mutagenic potential of chemical substances. Ames test involves use of bacteria called Salmonella typhimurium that grows well on agar with minimal nutrition requirements and is able to synthesize all amino acids. Ames developed a strain of S. typhimurium with mutated gene for synthesis of amino acid histidine. The histidine has to be added into the media otherwise this strain cannot survive. The principle of Ames test is based on the hypothesis that the application of mutagen leads mutations in many genes including the defective gene and some of those mutations cause the reversal of ability to synthesize histidine (reverse mutations). After subjection of his- S. typhimurium to the tested substance, the bacteria are placed on agar that does not contain histidine. After the incubation, the ratio is calculated between the surviving control bacteria not exposed to mutagen (the survival is caused by spontaneous mutation) and the surviving experimental bacteria that were exposed to mutagen. If there are many surviving bacteria within the experimental (=exposed group), it is considered as a proof of mutagenicity of the tested substance.
Arabidopsis thaliana
Probably the most important plant genetic model, the first plant with a completed genome sequence.
Eugenics
The term eugenics was first used by British mathematician and scientist, Sir Francis Galton. Eugenics studies methods that should lead to the best human genetic pool. Even though Plato defined the basic principle of positive eugenics - that the superior offspring comes from mating of the "best" men and women, the eugenics influenced the human history mainly due to its negative form in the ideology of Nazi Germany (in eugenics name many millions of people of "undesirable" race or health characteristics were killed), but also in sterilization of mentally ill people, epileptics and prisoners as bearers of "defective" and undesirable heritable traits in USA within years 1902 and 1964 (approximately 63 000 sterilizations). Ethical questions in eugenics context arise nowadays mainly due to in vitro fertilization and preimplantation genetic embryo testing.
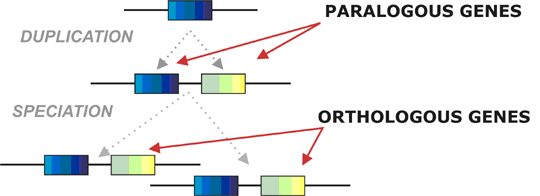
Homologous genes
Homologous genes are genes derived from a single ancestral gene. In 1970 it was first suggested to divide homologous genes into two types: paralogous and orthologous. Paralogous genes arise from ancestral gene duplication while the orthologous genes are products of speciation. There are numerous examples of both classes, e.g. human apolipoprotein genes are paralogous while the genes coding for apolipoprotein B in human and rat are orthologous.
MicroRNA
The group of small RNA called microRNA (miRNA) was only recently identified in the genomes of plants and animals. MiRNAs are an evolutionary conserved group that functions in fundamental biological processes. Thanks to their specific traits and predicted function the miRNAs are now on top of scientific interest. "Mature" molecules of miRNA are app. 22 nucleotides long products of cleavage of the inactive precursor. Nowadays there are about 1000 miRNAs identified, about one quarter of those in humans. It is presumed that genes for miRNAs number about 1% of all predicted genes. The amount of miRNA molecules in the cells dramatically differs according to the type of the cells, the ontogenetic stage - from a few copies up to 50,000. Similarly to mRNA, the amount of miRNA is thought to correspond to the miRNA activity in a given context. MicroRNA are not translated to proteins but directly act in regulation (repression) of target mRNA transcription, probably on the basis of complementarity of sequences of molecules. This newly found mechanism of post-transcription regulation fundamentally corrected the view on the process of realization of genetic information. MicroRNAs were shown to be important regulators of many physiological processes, e.g. in hematopoesis, insulin secretion, the nervous system or cancerogenesis. The knowledge that organisms naturally use short RNA molecules for gene expression regulation led to the development of RNA interference techniques (see below).
RNA interference
It's a technique based on recent finding that plants and animals naturally use short RNA molecules for gene expression regulation. Thanks to this technique it is feasible to investigate the function of genes, signaling and metabolic pathways, test the effect of drugs and also create so called knock-down models. Via application of short double stranded RNAs that do not code any protein, the silencing of target gene expression based on sequence complementarity of transferred dsRNA and endogenously transcribed mRNA is acquired. Two types of dsRNA that are used for those purposes, are called short interfering RNAs (siRNA) and short hairpin RNA (shRNA). After siRNA or shRNA introgression the target gene expression is assessed to find out the level of its silencing.
SNP
SNP (single nucleotide polymorphism). In 99.9%, people do not differ in their DNA sequence. Out of the remaining 0.1%, SNPs represent 80%. The Human Genome Project is being complemented by identification of millions of SNPs, made available by public databases, like dbSNP. The possibility of genotyping of 500,000 + in a single DNA sample should facilitate identification of alleles responsible for prevalent human diseases.